|
GLYLIB
0.3.0b
|
|
GLYLIB
0.3.0b
|
#include <molecules.h>
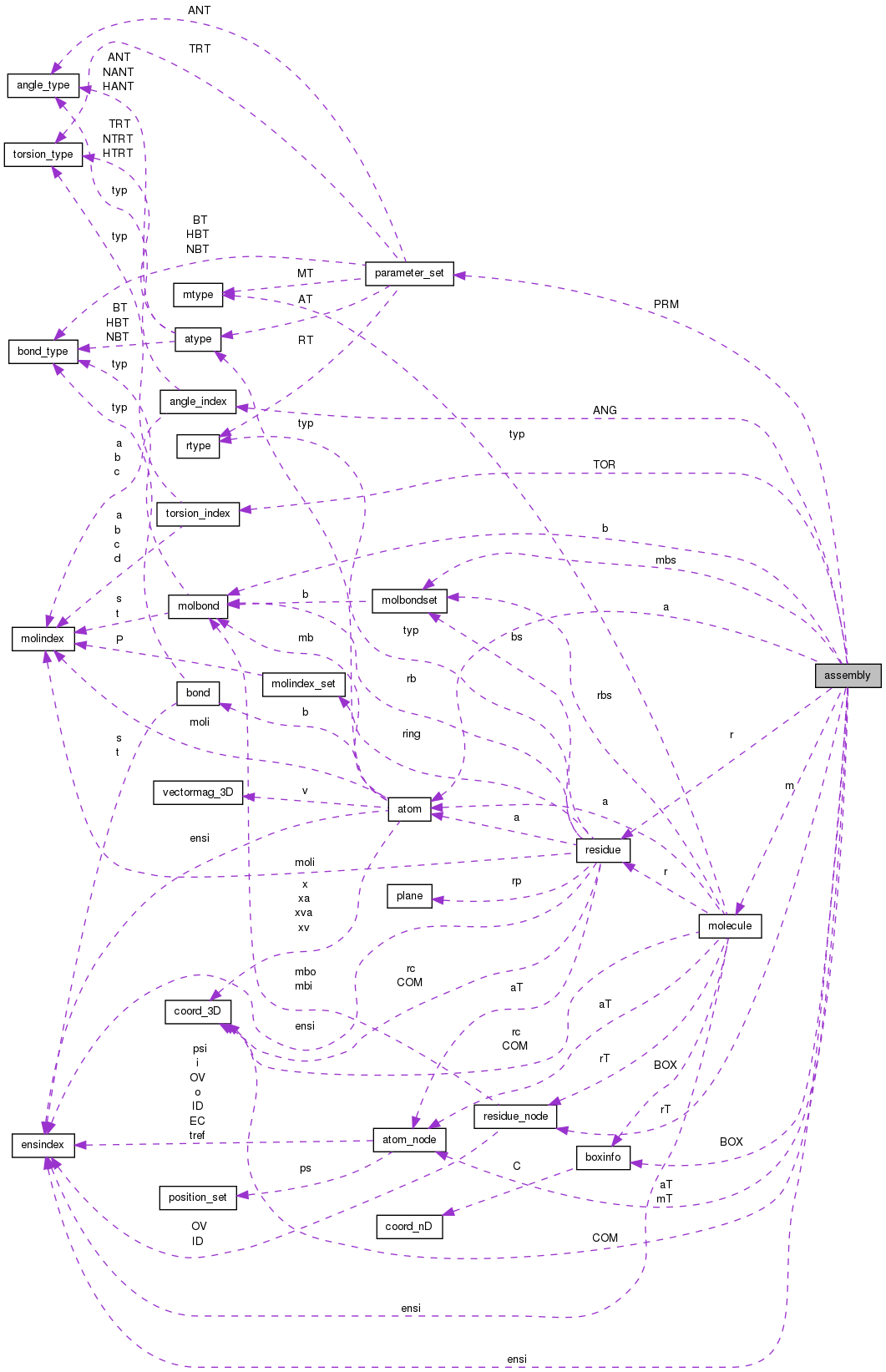
Public Attributes | |
| int | i |
| index | |
| char * | N |
| name | |
| char * | T |
| type | |
| char * | D |
| description (free-form) (was *desc) | |
| double | mass |
| mass of assembly | |
| double | TIME |
| current/initial/reference time in the simulation | |
| coord_3D | COM |
| center of mass | |
| int | nm |
| number of molecule structures | |
| molecule ** | m |
| nm of these | |
| atom_node * | mT |
| nm of these | |
| int | nr |
| number of residues | |
| residue ** | r |
| nr of these -- probably best to point into molecules... | |
| residue_node * | rT |
| nr of these | |
| int | na |
| number of atoms | |
| atom ** | a |
| na of these -- probably best to point into mol/res combos | |
| atom_node * | aT |
| na of these | |
| int | nb |
| total number of bonds | |
| molbond * | b |
| the bonds | |
| int | nANG |
| # of angles | |
| angle_index * | ANG |
| nANG of these | |
| int | nTOR |
| # of torsions | |
| torsion_index * | TOR |
| nTOR of these | |
| int | nPRM |
| number of parameter sets | |
| parameter_set ** | PRM |
| pointers to parameter sets | |
| int | nmbs |
| number of sets of connections between molecules | |
| molbondset * | mbs |
| nmbs of these sets | |
| int | nBOX |
| Number of box_info structures defined. | |
| boxinfo * | BOX |
| The nBOX structures. | |
| int | nOD |
| number of other descriptors | |
| char ** | OD |
| the nOD descriptors | |
| int | nensi |
| number of ensemble indices | |
| ensindex * | ensi |
| list of ensemble indices | |
| int | nVP |
| number of void pointers | |
| void * | VP |
| void pointers | |
Definition at line 289 of file molecules.h.
na of these -- probably best to point into mol/res combos
Definition at line 306 of file molecules.h.
Referenced by add_assembly_to_ensemble(), add_trajcrds_to_prmtop_assembly(), find_assembly_top_level_atoms_by_n(), get_moiety_selection_assembly(), getAssembly(), parse_amber_prmtop(), and set_assembly_atom_molbonds_from_PDB_CONECT().
na of these
Definition at line 308 of file molecules.h.
the bonds
Definition at line 310 of file molecules.h.
Referenced by initialize_assembly(), and parse_amber_prmtop().
The nBOX structures.
Definition at line 320 of file molecules.h.
Referenced by add_trajcrds_to_prmtop_assembly(), and parse_amber_prmtop().
center of mass
Definition at line 296 of file molecules.h.
Referenced by initialize_assembly(), and set_assembly_molecule_COM().
| char* assembly::D |
description (free-form) (was *desc)
Definition at line 293 of file molecules.h.
Referenced by parse_amber_prmtop().
list of ensemble indices
Definition at line 327 of file molecules.h.
| int assembly::i |
nm of these
Definition at line 298 of file molecules.h.
Referenced by add_assembly_to_ensemble(), get_assembly_molecule_COM(), get_assembly_PDB_ATOM_lines(), get_moiety_selection_assembly(), getAssembly(), initialize_assembly(), load_amber_prmtop(), parse_amber_prmtop(), and set_assembly_molecule_COM().
| double assembly::mass |
mass of assembly
Definition at line 294 of file molecules.h.
Referenced by get_assembly_molecule_COM(), initialize_assembly(), and set_assembly_molecule_COM().
nm of these
Definition at line 299 of file molecules.h.
| char* assembly::N |
name
Definition at line 291 of file molecules.h.
| int assembly::na |
number of atoms
Definition at line 305 of file molecules.h.
Referenced by add_assembly_to_ensemble(), add_trajcrds_to_prmtop_assembly(), find_assembly_top_level_atoms_by_n(), get_moiety_selection_assembly(), parse_amber_prmtop(), and set_assembly_atom_molbonds_from_PDB_CONECT().
| int assembly::nANG |
# of angles
Definition at line 311 of file molecules.h.
Referenced by deallocateAssembly(), and parse_amber_prmtop().
| int assembly::nb |
total number of bonds
Definition at line 309 of file molecules.h.
Referenced by deallocateAssembly(), initialize_assembly(), and parse_amber_prmtop().
| int assembly::nBOX |
Number of box_info structures defined.
Definition at line 319 of file molecules.h.
Referenced by add_trajcrds_to_prmtop_assembly(), deallocateAssembly(), initialize_assembly(), and parse_amber_prmtop().
| int assembly::nensi |
number of ensemble indices
Definition at line 326 of file molecules.h.
| int assembly::nm |
number of molecule structures
Definition at line 297 of file molecules.h.
Referenced by add_assembly_to_ensemble(), deallocateFullAssembly(), get_assembly_molecule_COM(), get_assembly_PDB_ATOM_lines(), get_moiety_selection_assembly(), getAssembly(), initialize_assembly(), load_amber_prmtop(), outputAsmblPDB(), parse_amber_prmtop(), set_assembly_molecule_COM(), and set_assembly_molindexes().
| int assembly::nmbs |
number of sets of connections between molecules
Definition at line 317 of file molecules.h.
Referenced by deallocateAssembly(), and initialize_assembly().
| int assembly::nOD |
number of other descriptors
Definition at line 324 of file molecules.h.
Referenced by deallocateAssembly().
| int assembly::nPRM |
number of parameter sets
Definition at line 315 of file molecules.h.
Referenced by deallocateAssembly(), and parse_amber_prmtop().
| int assembly::nr |
number of residues
Definition at line 302 of file molecules.h.
Referenced by add_assembly_to_ensemble(), get_moiety_selection_assembly(), and parse_amber_prmtop().
| int assembly::nTOR |
# of torsions
Definition at line 313 of file molecules.h.
Referenced by deallocateAssembly(), and parse_amber_prmtop().
| int assembly::nVP |
number of void pointers
Definition at line 328 of file molecules.h.
Referenced by initialize_assembly(), and parse_amber_prmtop().
| char** assembly::OD |
the nOD descriptors
Definition at line 325 of file molecules.h.
pointers to parameter sets
Definition at line 316 of file molecules.h.
Referenced by parse_amber_prmtop().
nr of these -- probably best to point into molecules...
Definition at line 303 of file molecules.h.
Referenced by add_assembly_to_ensemble(), get_moiety_selection_assembly(), and parse_amber_prmtop().
nr of these
Definition at line 304 of file molecules.h.
| char* assembly::T |
type
Definition at line 292 of file molecules.h.
| double assembly::TIME |
current/initial/reference time in the simulation
Definition at line 295 of file molecules.h.
| void* assembly::VP |