|
GLYLIB
0.3.0b
|
|
GLYLIB
0.3.0b
|
#include <molecules.h>
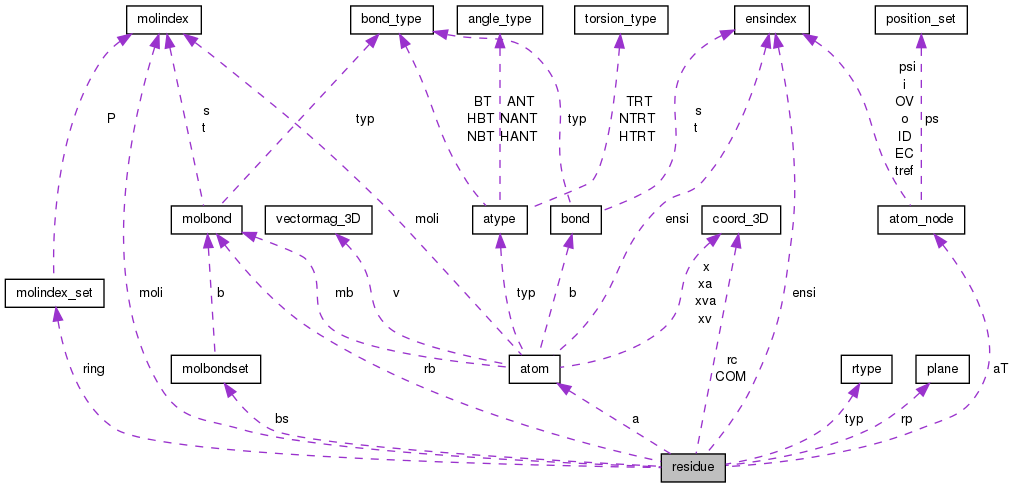
Public Attributes | |
| int | n |
| residue number given in input file | |
| char * | cID |
| char * | IC |
| insertion code | |
| char * | N |
| residue name | |
| char * | T |
| residue type | |
| char * | D |
| free-form description for residue | |
| int | t |
| type number | |
| rtype * | typ |
| pointer to rtype structure | |
| molindex | moli |
| the molecule index for this residue (address -- set .a to -1 or 0) | |
| int | na |
| number of atoms in residue | |
| atom * | a |
| atom structures (na of these) | |
| atom_node * | aT |
| na of these (one per atom) | |
| double | m |
| molecular weight | |
| coord_3D | COM |
| center of mass for residue | |
| int | nrb |
| number of bonds between residues | |
| molbond * | rb |
| nrb of these descriptions of bonds | |
| int | mTi |
| Index into the molecule's connection tree for this residue (m.rT) | |
| int | nbs |
| number of bond sets | |
| molbondset * | bs |
| (consecutive bonds, use these for plotting, etc.) | |
| int | nring |
| number of simple rings (no cage structures, etc.) | |
| molindex_set * | ring |
| molecule indices for the nring rings | |
| int | nrc |
| coord_3D * | rc |
| coordinates for ring/reference centers | |
| int | nrp |
| number of ring planes defined | |
| plane * | rp |
| equations for average/approximate/exact/etc. ring planes (where useful) | |
| int | ni |
| number of other indices | |
| int * | i |
| other indices, as needed (ni of these) | |
| int | nd |
| number of double-precision parameters | |
| double * | d |
| other parameters, as needed (nd of these) | |
| int | nensi |
| number of ensemble indices | |
| ensindex * | ensi |
| list of ensemble indices | |
| char * | altname |
| name of some other/original name for this residue | |
| int | nOD |
| number of other descriptors | |
| char ** | OD |
| the nOD descriptors | |
| int | nVP |
| number of void structures | |
| void * | VP |
| nVP of these, whatever they may be | |
Definition at line 180 of file molecules.h.
atom structures (na of these)
Definition at line 191 of file molecules.h.
Referenced by add_assembly_to_ensemble(), find_residue_atoms_by_N(), follow_molecule_atom_molbonds_for_contree(), follow_molecule_atom_nodes_from_bonds(), follow_residue_atom_nodes_from_bonds(), get_assembly_molecule_COM(), get_assembly_PDB_ATOM_lines(), get_ensemble_COM(), get_ensemble_PDB_ATOM_lines(), get_moiety_selection_assembly(), get_molecule_COM(), get_molecule_PDB_ATOM_lines(), get_molecule_point_charge_dipole(), get_residue_COM(), get_residue_PDB_ATOM_lines(), initialize_residue(), load_dlg_mol(), normalize_molecule_vectors(), parse_amber_prmtop(), read_prepres(), rotate_vector_to_Z_M(), set_connection_tree_molecule_atoms(), set_molecule_atom_nodes_from_bonds(), set_molecule_residue_molbonds(), set_residue_atom_nodes_from_bonds(), set_residue_molindexes(), shift_molecule_atoms_by_vector_scale(), translate_ensemble_by_XYZ(), translate_molecule_by_XYZ(), translate_residue_by_XYZ(), and translate_zero_to_coord_M().
| char* residue::altname |
name of some other/original name for this residue
Definition at line 215 of file molecules.h.
Referenced by initialize_residue().
na of these (one per atom)
Definition at line 192 of file molecules.h.
Referenced by follow_residue_atom_nodes_from_bonds(), initialize_residue(), and set_residue_atom_nodes_from_bonds().
(consecutive bonds, use these for plotting, etc.)
Definition at line 199 of file molecules.h.
Referenced by initialize_residue(), and read_prepres().
| char* residue::cID |
Chain identifier
Definition at line 182 of file molecules.h.
Referenced by getResInfo(), and initialize_residue().
center of mass for residue
Definition at line 194 of file molecules.h.
Referenced by initialize_residue(), load_dlg_mol(), read_prepres(), set_molecule_COM(), and set_residue_COM().
| char* residue::D |
free-form description for residue
Definition at line 186 of file molecules.h.
Referenced by initialize_residue(), and read_prepres().
| double* residue::d |
other parameters, as needed (nd of these)
Definition at line 212 of file molecules.h.
Referenced by initialize_residue(), and read_prepres().
list of ensemble indices
Definition at line 214 of file molecules.h.
Referenced by parse_amber_prmtop().
| int* residue::i |
other indices, as needed (ni of these)
Definition at line 210 of file molecules.h.
Referenced by get_assembly_PDB_ATOM_lines(), get_ensemble_PDB_ATOM_lines(), get_residue_PDB_ATOM_lines(), initialize_residue(), and read_prepres().
| char* residue::IC |
insertion code
Definition at line 183 of file molecules.h.
Referenced by getMolecule(), getResInfo(), and initialize_residue().
| double residue::m |
molecular weight
Definition at line 193 of file molecules.h.
Referenced by follow_molecule_residue_nodes_from_bonds(), get_residue_COM(), initialize_residue(), load_dlg_mol(), read_prepres(), and set_molecule_COM().
the molecule index for this residue (address -- set .a to -1 or 0)
Definition at line 189 of file molecules.h.
Referenced by add_assembly_to_ensemble(), parse_amber_prmtop(), and set_residue_molindexes().
| int residue::mTi |
Index into the molecule's connection tree for this residue (m.rT)
Definition at line 197 of file molecules.h.
Referenced by follow_molecule_residue_nodes_from_bonds(), initialize_residue(), and set_molecule_residue_nodes_from_bonds().
| int residue::n |
residue number given in input file
Definition at line 181 of file molecules.h.
Referenced by get_molecule_PDB_ATOM_lines(), get_PDB_line_for_ATOM(), get_residue_PDB_ATOM_lines(), initialize_residue(), parse_amber_prmtop(), and read_prepres().
| char* residue::N |
residue name
Definition at line 184 of file molecules.h.
Referenced by getResInfo(), initialize_residue(), parse_amber_prmtop(), and read_prepres().
| int residue::na |
number of atoms in residue
Definition at line 190 of file molecules.h.
Referenced by add_assembly_to_ensemble(), deallocateResidue(), find_residue_atoms_by_N(), follow_residue_atom_nodes_from_bonds(), get_alt_rms_mol(), get_alt_rms_res(), get_assembly_molecule_COM(), get_assembly_PDB_ATOM_lines(), get_ensemble_COM(), get_ensemble_PDB_ATOM_lines(), get_moiety_selection_assembly(), get_molecule_COM(), get_molecule_PDB_ATOM_lines(), get_molecule_point_charge_dipole(), get_residue_COM(), get_residue_PDB_ATOM_lines(), initialize_residue(), load_dlg_mol(), normalize_molecule_vectors(), parse_amber_prmtop(), read_prepres(), rotate_vector_to_Z_M(), set_connection_tree_molecule_atoms(), set_molecule_atom_nodes_from_bonds(), set_molecule_residue_molbonds(), set_residue_atom_nodes_from_bonds(), set_residue_molindexes(), shift_molecule_atoms_by_vector_scale(), translate_ensemble_by_XYZ(), translate_molecule_by_XYZ(), translate_residue_by_XYZ(), and translate_zero_to_coord_M().
| int residue::nbs |
number of bond sets
Definition at line 198 of file molecules.h.
Referenced by deallocateResidue(), initialize_residue(), and read_prepres().
| int residue::nd |
number of double-precision parameters
Definition at line 211 of file molecules.h.
Referenced by initialize_residue(), and read_prepres().
| int residue::nensi |
number of ensemble indices
Definition at line 213 of file molecules.h.
Referenced by parse_amber_prmtop().
| int residue::ni |
number of other indices
Definition at line 209 of file molecules.h.
Referenced by get_assembly_PDB_ATOM_lines(), get_ensemble_PDB_ATOM_lines(), initialize_residue(), and read_prepres().
| int residue::nOD |
number of other descriptors
Definition at line 216 of file molecules.h.
Referenced by deallocateResidue(), and initialize_residue().
| int residue::nrb |
number of bonds between residues
Definition at line 195 of file molecules.h.
Referenced by follow_molecule_residue_nodes_from_bonds(), initialize_residue(), and set_molecule_residue_molbonds().
| int residue::nrc |
Definition at line 204 of file molecules.h.
Referenced by initialize_residue(), and load_dlg_mol().
| int residue::nring |
number of simple rings (no cage structures, etc.)
Definition at line 200 of file molecules.h.
Referenced by initialize_residue().
| int residue::nrp |
number of ring planes defined
Definition at line 206 of file molecules.h.
Referenced by initialize_residue().
| int residue::nVP |
number of void structures
Definition at line 218 of file molecules.h.
Referenced by initialize_residue(), and read_prepres().
| char** residue::OD |
nrb of these descriptions of bonds
Definition at line 196 of file molecules.h.
Referenced by follow_molecule_residue_nodes_from_bonds(), initialize_residue(), and set_molecule_residue_molbonds().
coordinates for ring/reference centers
Definition at line 205 of file molecules.h.
Referenced by initialize_residue(), and load_dlg_mol().
molecule indices for the nring rings
Definition at line 201 of file molecules.h.
equations for average/approximate/exact/etc. ring planes (where useful)
Definition at line 207 of file molecules.h.
Referenced by initialize_residue().
| char* residue::T |
| int residue::t |
pointer to rtype structure
Definition at line 188 of file molecules.h.
Referenced by deallocateResidue(), and initialize_residue().
| void* residue::VP |
nVP of these, whatever they may be
Definition at line 219 of file molecules.h.
Referenced by read_prepres().