Contents
Overview
Learn how to predict which glycans are likely to bind to a given protein.
To do this, we will use the Gly-Spec Webtool, which uses the 3D structure of a glycan-binding protein in complex with a carbohydrate fragment that is a minimal binding determinant (MBD) for that protein. The webtool finds glycans that contain the MBD within their structure, and predicts if they will bind to the protein.
Example System
This scenario can be followed with a co-complex of your choice, but if you don’t have one, the following files have what you need to follow along:
The zip file contains a pdb file of the UDA lectin (Urtica dioica agglutinin), along with a glycan fragment that has been trimmed down to only the MBD. It also contains a simple text file which is a list of the glycan IDs of several glycans that are known to bind to UDA.
Background
The Gly-Spec tool was originally named the Grafting tool, and it was developed by Oliver Grant. The term Gly-Spec refers to the specificity of glycans.
What the tool does:
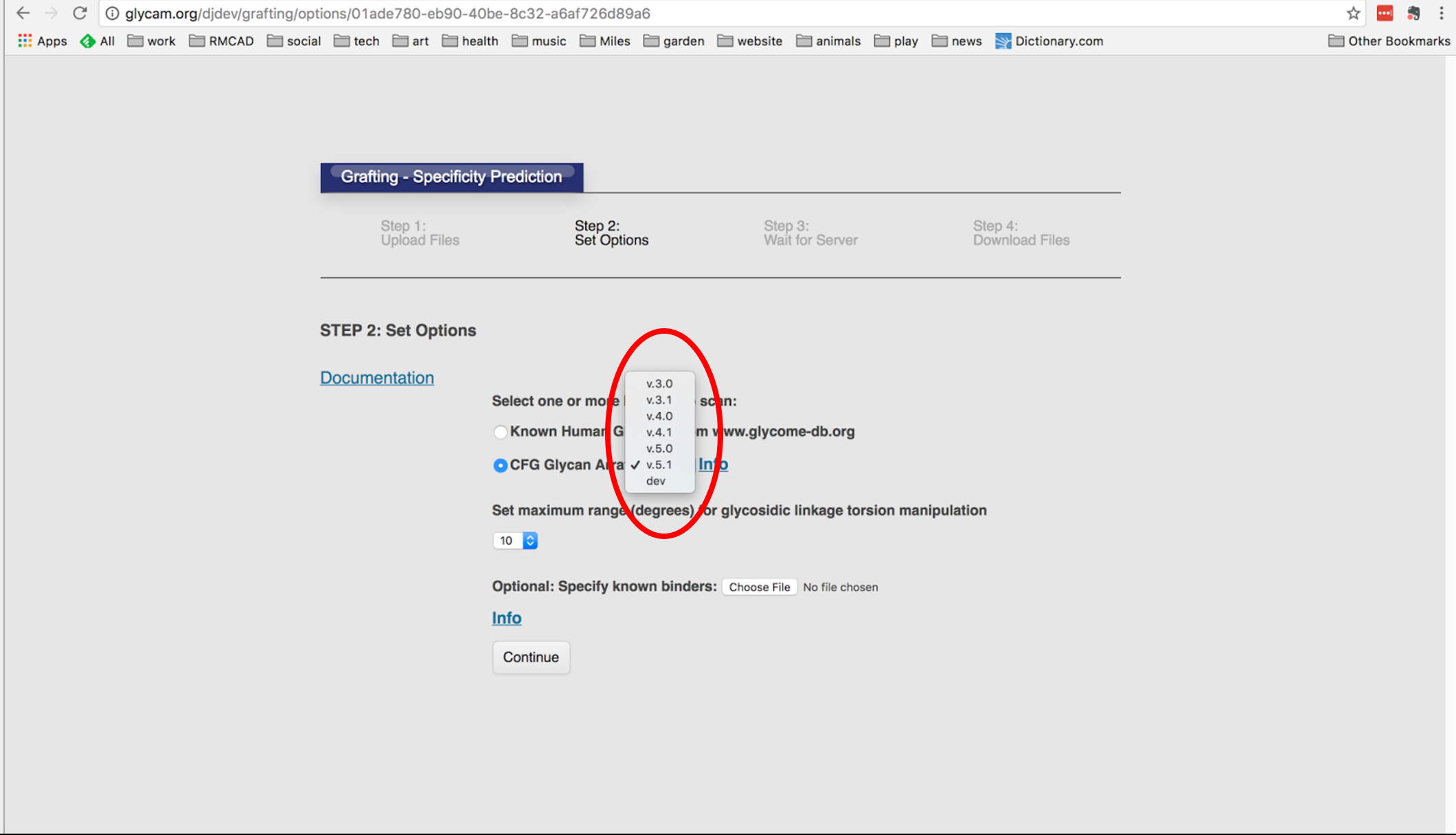
The Gly-Spec tool takes a protein in complex with a glycan fragment representing the MBD and scans a user-selected glycan array, or the human glycome, for glycans that contain the MBD.
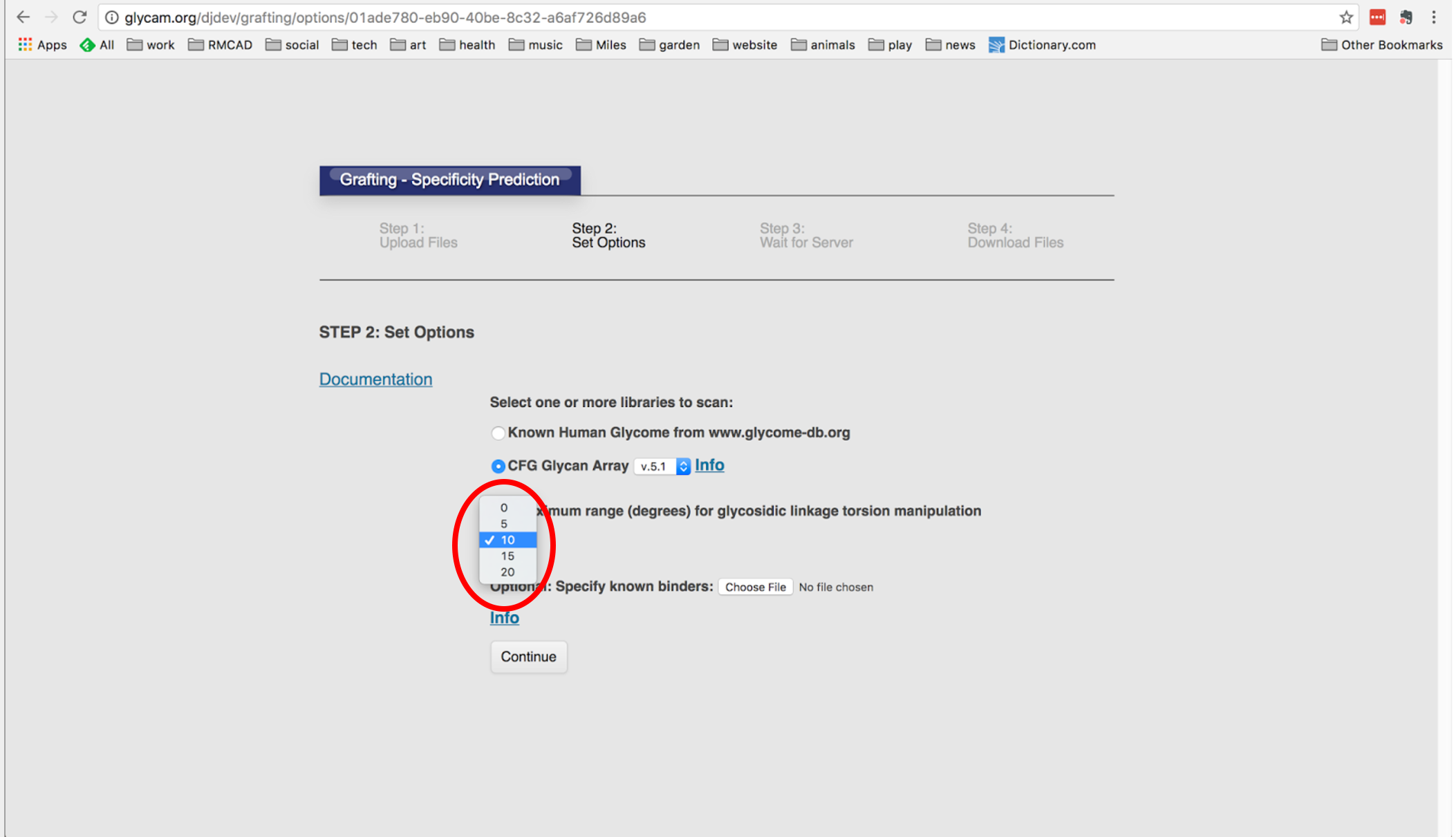
The Gly-Spec tool then tries to build each of the glycans it found into the binding site of the protein, by growing out the larger glycan from the co-complexed MBD. If the attached “branches” have any atomic overlaps with the protein binding site the webtool attempts to resolve them by “wiggling” the interglycosidic linkages within limits that can be set by the user. In cases where the adjustments do not resolve the overlap, the tool predicts that the glycans will not bind. In cases where all overlaps are resolved by wiggling, the tool predicts the glycan will bind.
The Gly-Spec tool also considers whether the linkers used on a CFG glycan array might prevent binding of a glycan that otherwise would bind, and reports this to the user.
Finally, the Gly-Spec tool produces a report that lists predicted binders and non-binders, compares to any provided list of binders, and gives the user the opportunity to download the 3D structure files for the glycans that were screened. This includes both the predicted binders and non-binders. These can be viewed in programs like VMD, to visually demonstrate the structure of binders in complex with your protein, or to assess the particular protein residues or structural features that prevent certain glycans from binding.
Required Input
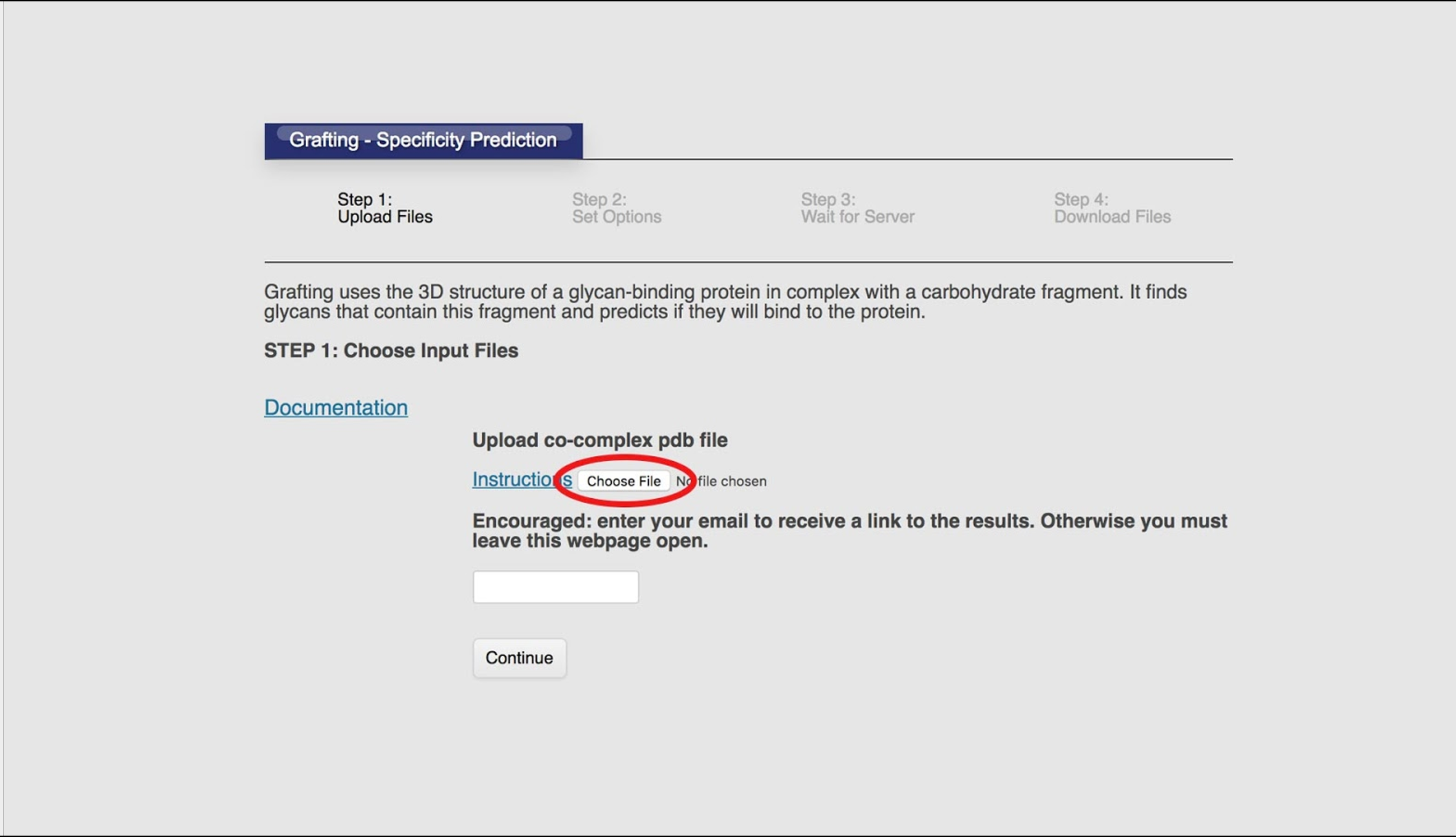
The tool requires a pdb file of a protein and a glycan fragment in complex. If you don’t have a co-complex, and just want to learn, see the Example System section above.
Optionally, we can also upload a text file containing a list of the glycan ids of known binders. Most often you want to, because this allows the tool to estimate its accuracy. Known binders are usually determined as the result of experimental screening data.
Protocol
Annotated Output
The 3D structures are provided in the form of PDB files. These can be viewed in modeling applications, such as VMD.
Additional Info
In case someone wants additional information about this or that.
Alternate Protocols
We are still developing the alternate protocols.
Troubleshooting
Tips for the user.