Return to Grafting documentation | Tutorials | Instructions
Tutorial 2 – Discovering Hidden Minimal Binding Determinants in your Array Data
Click here to download Tutorial 2 Material
Background
The hemagglutinating activity of Arachis hypogaea (Peanut) Agglutinin (PNA) has been reported to be most potently inhibited by glycans containing Galβ1-3GalNAc (color is used throughout to help relate between the text and tables, but is not required). The disaccharides lactose (Galβ1-4Glc) and lactosamine (Galβ1-4GlcNAc) were shown to inhibit PNA hemagglutinating activity, with around 50 fold less potency than the Galβ1-3GalNAc disaccharide.
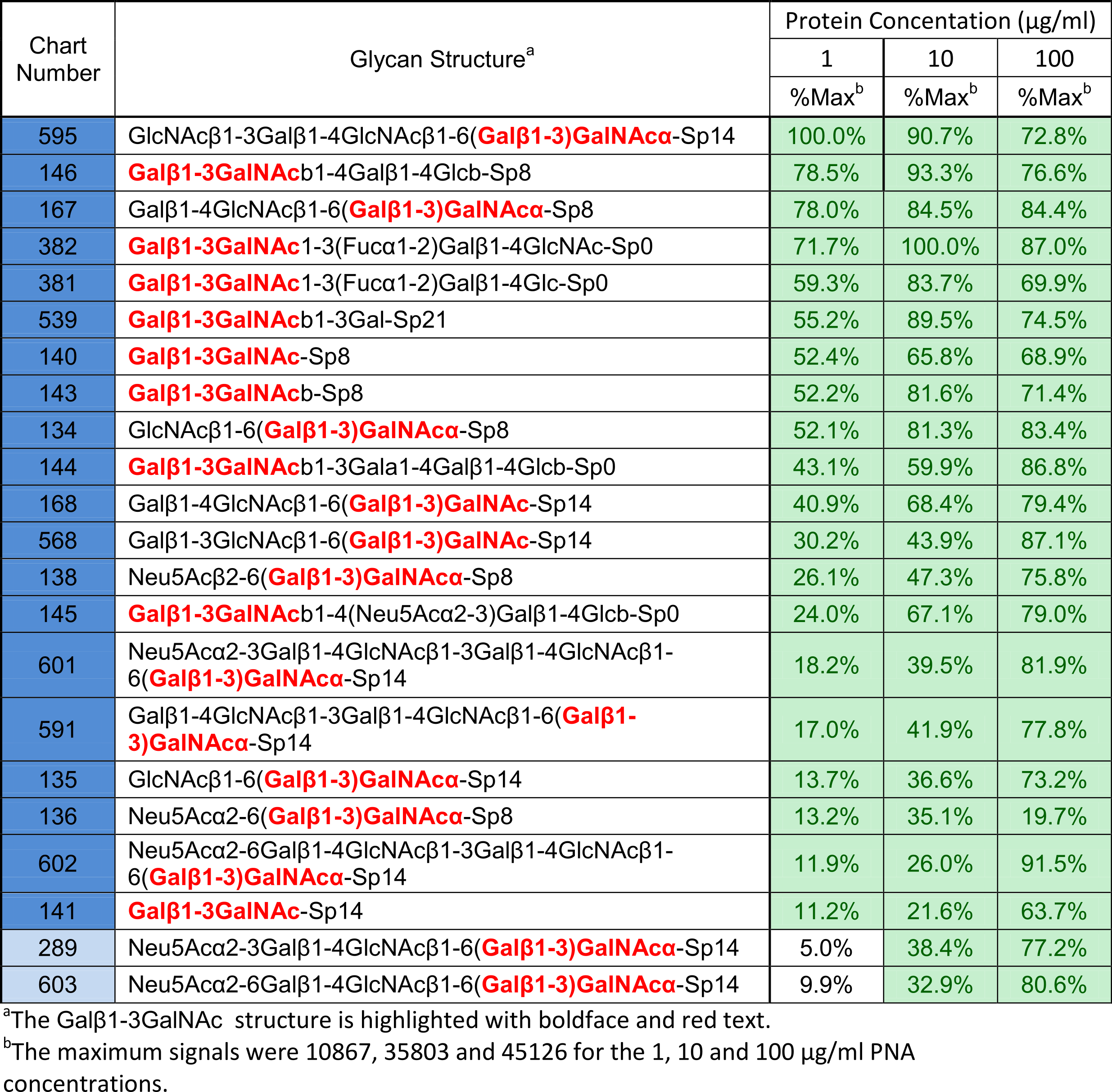
Glycan array screening data (CFG v5.0) shows that, as expected, Galβ1-3GalNAc containing structures were present in dominant binders. These glycans contain either Galβ1-3GalNAcα, which we are designating “minimal binding determinant 1″ (MBD1), or Galβ1-3GalNAcβ (MBD2). See Table 1. MBD1 is also known as the cancer associated TF-antigen.
Table 1: The top 22 glycans that bound to PNA on the v5.0 CFG glycan array. Click here to learn about our ranking system.
As you can see, these top binding glycans all contain MBD1, which matches what has already been reported in the literature for PNA binding.
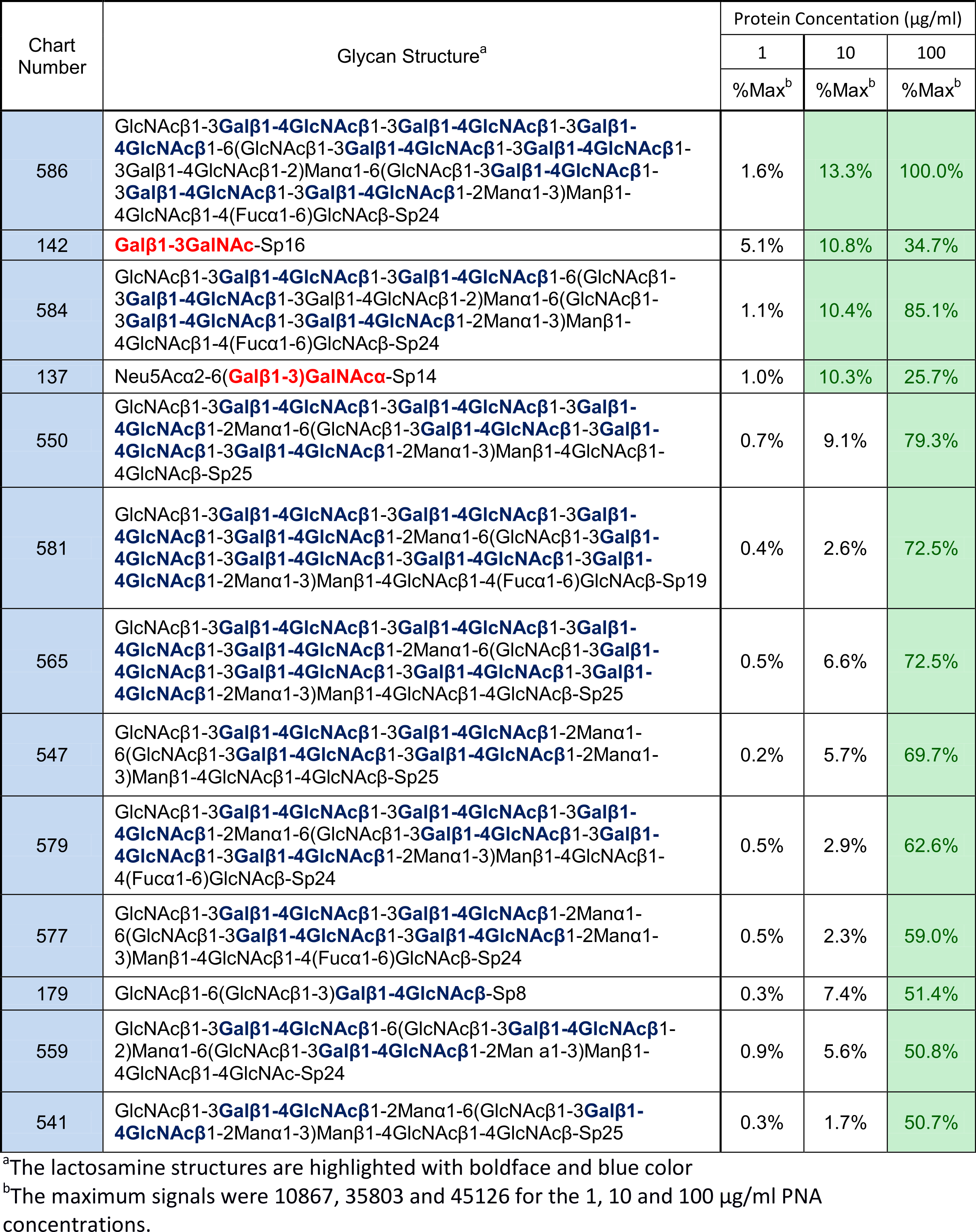
There are also a number of weak binders present that contain neither MBD1 nor MBD2, but do contain lactosamine. See Table 2.
Table 2: Glycans whose signals were categorized as “weak” for PNA binding
This also seems to match the literature, where lactosamine displayed weaker affinity than MBD1. Without any further investigation, it is natural to assume that lactosamine is also an MBD for PNA.
Step 1: Preform grafting with lactosamine against the CFG v5.0 by uploading the provided structure of PNA bound to lactosamine.
Step 2: Open the stats file for grafting lactosamine (called PNA-Lactosamine-Galb1-4GlcNAcb-stats.txt) and scroll down to the summary. You’ll see the lines:
Correctly predicted 205 non-binders out of 282 non-binders
Correctly predicted 3 binders out of 22 experimental binders
Overall this is a 43% agreement between the array data and the 3D structure. Something is definitely not correct. Most of the experimental binders do not fit in the binding site, while a large number of non-binders that contain lactosamine do fit in the binding site. This suggests that any affinity of lactose/lactosamine for PNA was insufficient for detection and it is not an MBD.
So how do we reconcile the data? What is causing the weak binding of these glycans?
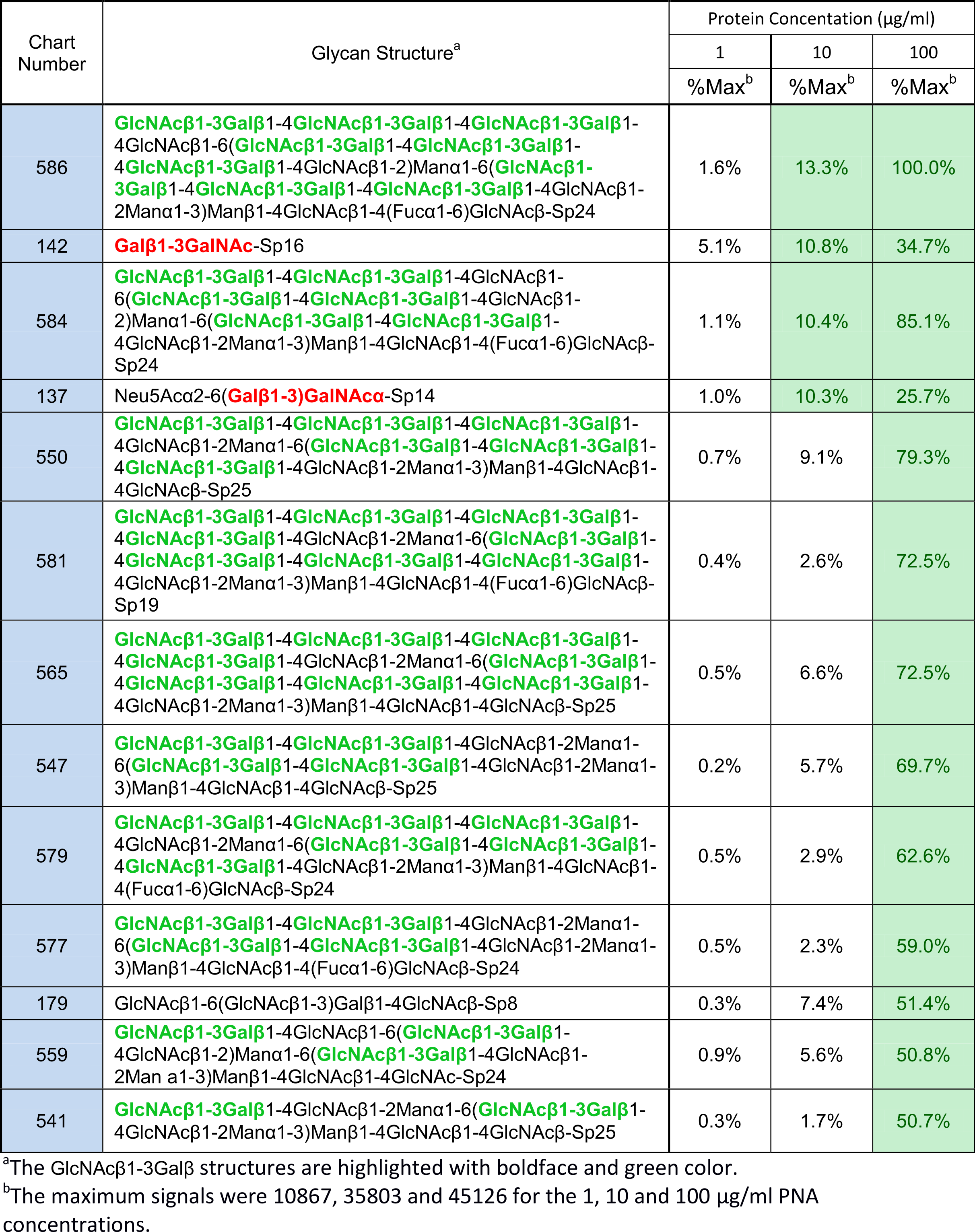
Let’s look at the data again, but with a different potential MBD highlighted (See Table 3).
Table 3: Glycans whose signals were categorized as “weak” for PNA binding
A previously unreported MBD comprised of GlcNAcβ1-3Galβ (MBD3) is present at the reducing termini of 13 weak binders (see Table 3), and a further 4 weak binders that also contain MBD1.
As an experimental 3D structure for MBD3 bound to PNA was unavailable, a model based on MBD1 was built. You can repeat the process in Step 3 if you wish to learn, or you can use the provided materials and skip to Step 4.
Step 3: If you wish to skip this step, MBD3 is included in the materials you downloaded at the beginning. Build a 3D structure of the proposed MBD GlcNAcβ1-3Galβ using the point-and-click interface of the carbohydrate builder on Glycam-Web; legacy.glycam.org/cb. Alternatively, you can build via this URL: http://legacy.glycam.org/url?condensed=DGlcpNAcb1-3DGalpb1-OH. Load this structure and MBD1 into UCSF Chimera, and superimpose the ring atoms onto those of MBD1 using the match command of Chimera. Delete MBD1 and combine the new MBD3 with the PNA structure. Save the PDB file.
Example of Chimera commands:
- match #1:0YB@C1,C3,C5 #0:0LB@C1,C3,C5
- delete #0:0LB,3VA,ROH
- combine #0,#1
Step 4: Perform grafting with GlcNAcβ1-3Galβ as the MBD. Follow the prompts at legacy.glycam.org/gr, remembering to select CFG v5.0 as the array. It will take around 30 minutes to complete and the stats files should be identical to those in the provided materials. Open the stats file (PNA-MBD3-GlcNAcb1-3Galb-stats.txt in the provided materials) and scroll down to the summary at the bottom. You will see these lines:
Correctly predicted 94 non-binders out of 109 non-binders
Correctly predicted 14 binders out of 17 experimental binders
Experimental Binders: Why only 14 out of 17? Well, there are 4 experimental binders (glycans 591, 595, 601 and 602) that contain both MBD1 and MBD3. Grafting predicts that 595 can bind via either motif, but the other three only bind via MBD1. Thus they show up as incorrectly predicted not to bind when using MBD3, but are correctly predicted to bind via MBD1.
Experimental Non-Binders: Why only 94 out of 109? We find that grafting results are not as accurate for weak binding motifs as for the dominant motifs, generally because we predict glycans to bind that do not show affinity on the array. We assume this is just the nature of the data and our use of arbitrary cut-offs, and we will not be able to reproduce 100% of the results.
Summary: When the results are combined using MBDs 1-3, grafting reproduces 90% of the array data for PNA.
Conclusion: The analysis suggests that PNA is less specific than previously thought, and provides a structural rationale for a new binding determinant (GlcNAcβ1-3Galβ, MBD3). This is particularly significant in light of the fact that PNA is used as a reagent thought to be able to selectively detect glycans which contain the cancer associated Galβ1-3GalNAcα (MBD1, TF-antigen) and Galβ1-3GalNAcβ (MBD2) motifs. Thus, the use of PNA as a reagent to exclusively detect Galβ1-3GalNAc-containing structures should be reconsidered, especially when studying biological samples, as avidity effects may be anticipated that would enhance binding to MBD3-containing glycans.