Contents
Description
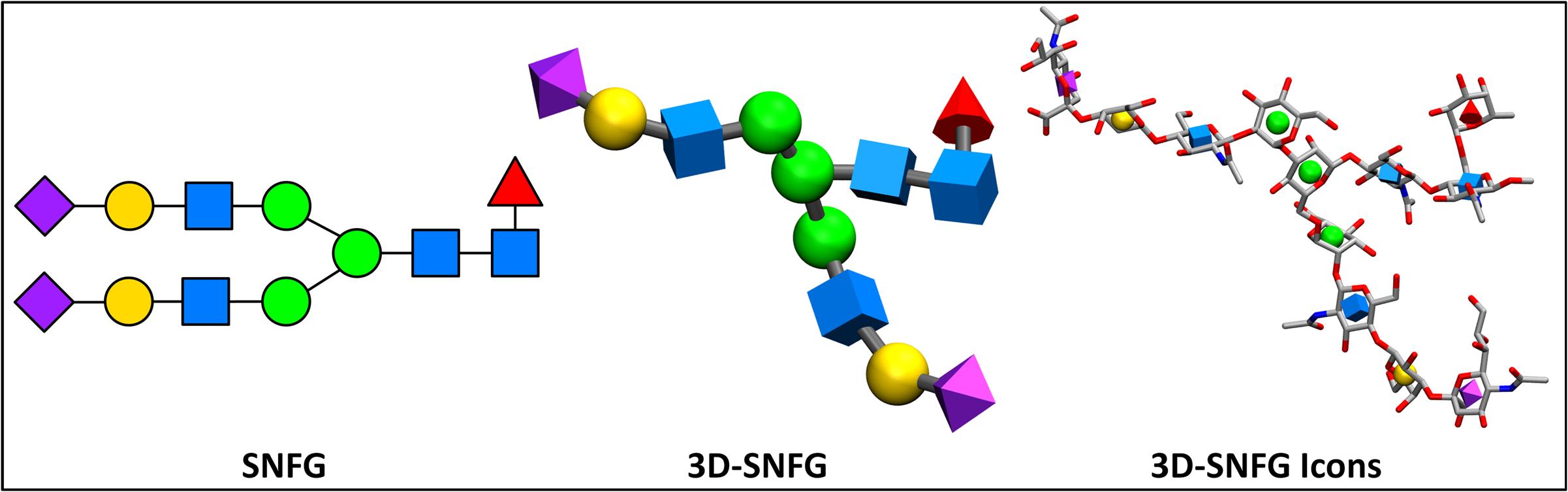
The Symbol Nomenclature For Glycans (SNFG) facilitates the efficient communication of carbohydrate structures, and has become widely accepted by the glycobiology community. The system is fully described in the NCBI text Essentials of Glycobiology.
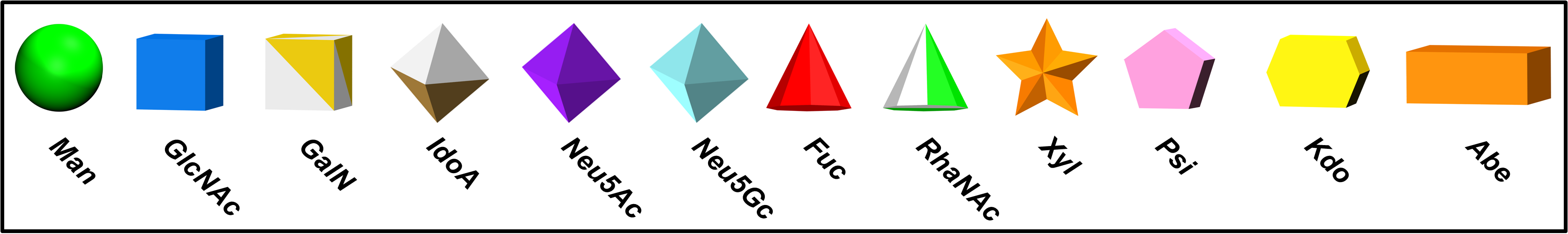
Carbohydrates currently lack a simplistic, standardized representation for 3D structure, and are thus predominantly visualized with atoms displayed. Discerning the glycan sequence from these atomistic views is challenging, even for short for oligosaccharide chains. The following software generates 3D shapes that match the SNFG format (both color and shape), and positions them on the geometric center of carbohydrate rings that are identified within a PDB file. This representation generates a cartoon-like representation for glycans within the Visual Molecular Dynamics (VMD) program. Automated recognition of monosaccharides relies upon commonly employed residue1 and atom names.2
1The current list of recognizable residue names includes common names from the PDB, as well those from the GLYCAM and CHARMM forcefields. Unknown residues are depicted as white, flat hexagons
2The ring atoms must be C1, C2, C3, C4, C5, and O5, unless the monosaccharide is a sialic acid, in which case they should be named C2, C3, C4, C5, C6, and O6
Installation
Note: VMD installation is discussed in the documentation of Ancilliary Software.
Step 1) Download
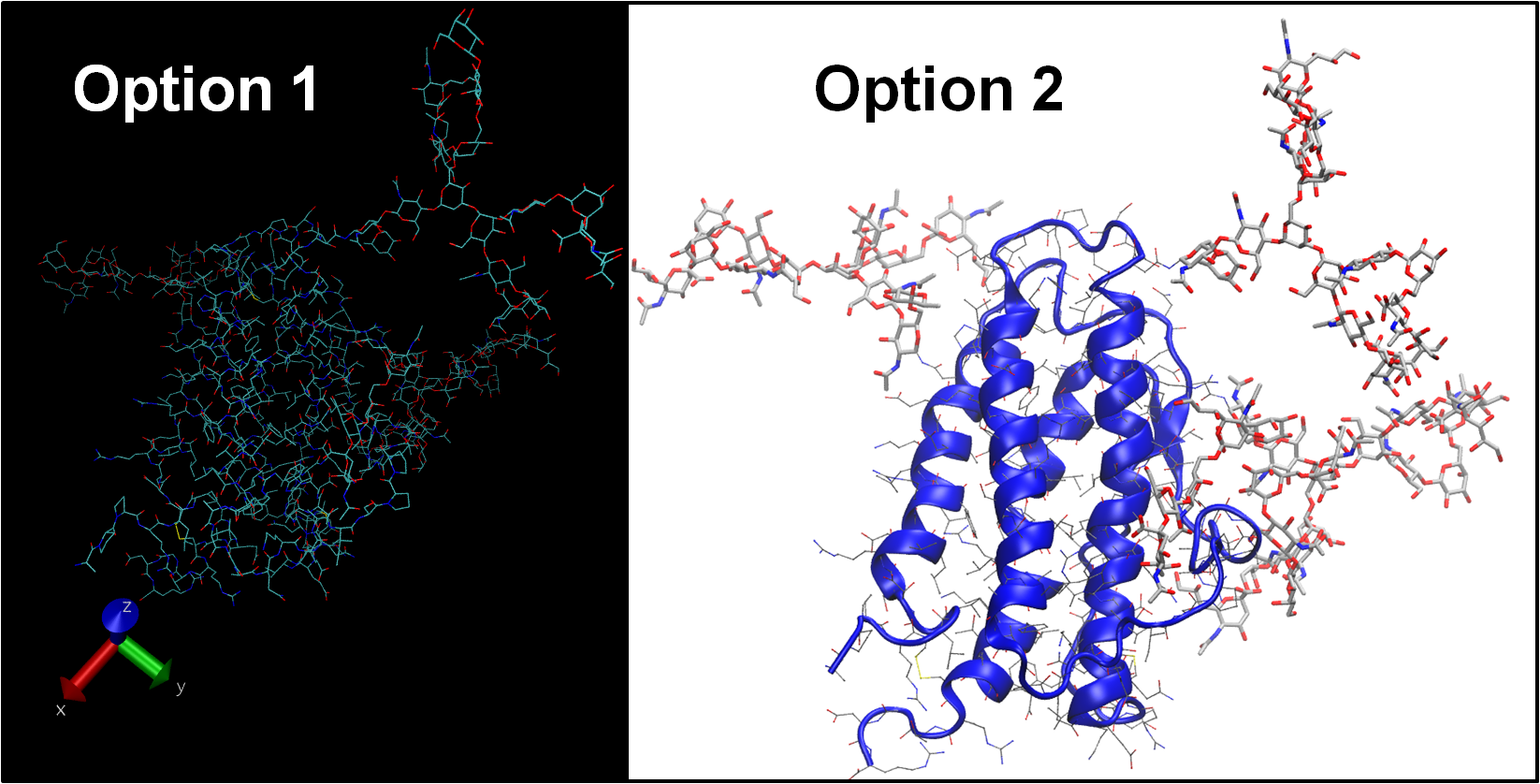
Two options are provided that differ by the VMD ‘default’ settings. Option 1 only includes the 3D-SNFG representations, and maintains the original settings for VMD. Option 2 contains the 3D-SNFG script, as well as a few aesthetic changes that are loaded at startup (i.e. white background, automatic representation of proteins with NewCartoon, etc.). The 3D-SNFG representations are identical in both cases.
Details: A vmdrc file allows you to customize the default settings of VMD. Option 2 initializes multiple representations in order to display the protein backbone as a cartoon, side chains as sticks, and all other atoms as licorice. The background has been changed to white, labels to black,, and display set to an orthographic view. Other options can be useful, such as changing the size and position of windows.
Option 1 | Option 2 |
|---|---|
| Linux & Macintosh: Download the following .vmdrc file Template file :: default does not exist! | Linux & Macintosh: Download the following .vmdrc file Template file :: default does not exist! |
| Windows: Download the following vmd.rc file Template file :: default does not exist! | Windows: Download the following vmd.rc file Template file :: default does not exist! |
Experienced VMD Users: You can source the 3D-SNFG script within your personal vmdrc file. The provided vmdrc files consist of two parts, the initialization of VMD and the 3D-SNFG tcl script. Rename the provided vmdrc file 3D-SNFG_v1.tcl, and remove all of the lines above ‘# Load the 3D-SNFG script’. Now source this script within your personal vmdrc.
Step 2) Install
Move the file to either your home directory, or the location where the VMD software is installed, and unzip the file.
Note that the unzipped file is named .vmdrc on Macintosh and Linux systems. A period that precedes a filename designates the file as “hidden”. Although the unzipped contents may appear empty, the file will be recognized by VMD.
Step 3) Visualize
Load a file containing a glycan into VMD (i.e. PDB ID: 3SGJ). On your keyboard, use the following shortcut keys:
- ‘i’ – apply the SNFG-Icons representation
- ‘g’ – apply the 3D-SNFG representation
- ‘b’ – apply the 3D-SNFG representation and label the reducing terminus
- ‘d’ – delete the drawn objects
FAQ
- Are structures available to test my setup?
- Does the code discriminate between D- and L- residues?
- How were the sizes of the 3D-SNFG shapes selected?
- How are modifications represented (i.e. sulfate and phosphate)?
- How do I draw 3D-SNFG objects on a molecule other than the most recently loaded file?
- How do I change the shape of an unrecognized residue (flat, white hexagon)?
- What does “ERROR) No molecules loaded” mean?
- Why are some of the shapes missing?
- Why are all of the shapes missing?
- How are the shapes built?
Advanced Usage
- Introduce new residue names
- Change the material of the 3D-SNFG shapes
- Color monosaccharide atoms according to the SNFG
- Combine 3D-SNFG with a Trajectory Smoothing Window
Gallery
Questions/Comments
If you have any issues that are not covered in the FAQ, or would like to provide suggestions for the representations, please contact us.
Citation
Thieker, D. F., Hadden, J. A., Schulten, K., & Woods, R. J. (2016). 3D implementation of the Symbol Nomenclature for Graphical Representation of Glycans. Glycobiology, 26(8), 786-787. DOI:10.1093/glycob/cww076)
Associated Publications
SNFG Update | Varki, A., Cummings, R. D., Aebi, M., Packer, N. H., Seeberger, P. H., Esko, J. D., … & Prestegard, J. J. (2015). “Symbol Nomenclature for Graphical Representations of Glycans.” Glycobiology, 25 (12), 1323-1324. DOI: 10.1093/glycob/cwv091
VMD | Humphrey, W., Dalke, A. & Schulten, K. (1996). “VMD – Visual Molecular Dynamics”, J. Molec. Graphics, 14 (1), 33-38. DOI: 10.1016/0263-7855(96)00018-5