Return to Grafting documentation | Tutorials | Instructions
Tutorial 1 – Screening a mAb against a glycan array
Click here to download Tutorial 1 Material
Background
JAA-F11 is a monoclonal antibody (mAb) that binds the TF-antigen (Galβ1-3GalNAcα). JAA-F11 was screened against a glycan array available from the consortium for functional glycomics (CFG). This tutorial demonstrates that grafting can accurately predict the specificity of JAA-F11 on the glycan array.
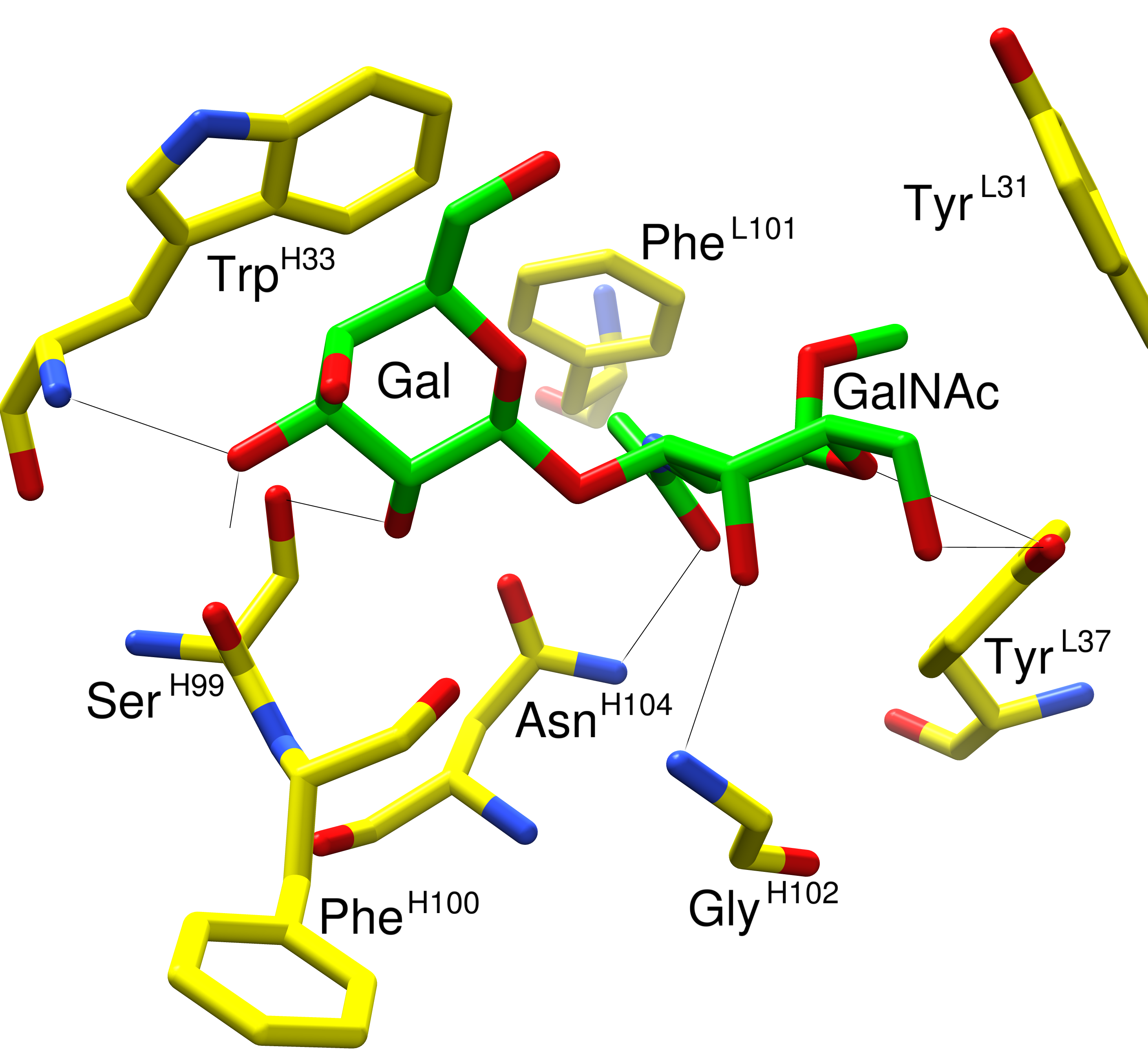
The tutorial material contains cocomplex.pdb and binders.txt. The cocomplex.pdb file contains a 3D structure of JAA-F11 in complex with Galβ1-3GalNAcα. You can visually inspect this file with software such as UCSF-Chimera, pymol or VMD. Here is an example that is focused on the interactions in the binding site:
The binders.txt contains a list of the IDs of glycans which JAA-F11 bound to on the CFG v4.0 glycan array. You can learn how I collate glycan array screening data and identify binders by clicking here. It is possible to view the resulting list of binders using a text-editor such as Wordpad in Windows. Here is what it should look like:
131
127
129
125
182
157
159
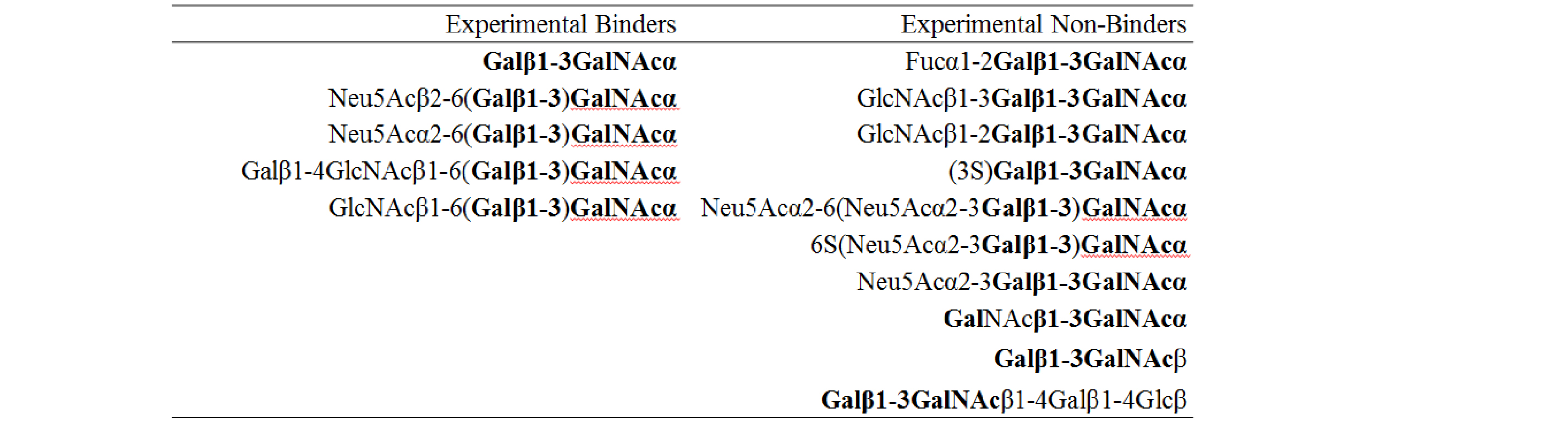
These numbers correspond to glycan ID numbers used on the CFG v4.0 array. They refer to a specific glycan, conjugated via a linker to the array surface. Each of the 442 glycans on v4.0 have a unique ID number. When JAA-F11 was screened against the array, it was observed to bind glycans with the above ID numbers. Some glycan structures are printed with different linkers. Linker effects will be addressed in a different tutorial, but for now we can conclude that JAA-F11 bound to five glycans that contained Galβ1-3GalNAcα. Interestingly, some glycans that contained Galβ1-3GalNAc in their structure were not bound by JAA-F11:
As 3D structure ultimately determines specificity, it should be possible to relate the experimentally observed specificity to the 3D structure JAA-F11 bound to Galβ1-3GalNAcα. I.e. the 6-position of the GalNAc must be open for substitution, whereas substitution at the 2- and 3-positions of the Gal prevent binding. Further, the Galβ1-3GalNAcβ structure is not bound by JAA-F11.
Open an internet browser and go to www.legacy.glycam.org/gr. Follow the instructions in the interface. Upload the cocomplex.pdb and experimental-binders.txt. Select array v4.0 and then wait for grafting to complete. It is possible to enter your email address and receive a link when the job completes. You can learn how to interpret the results by clicking here.
In this rather simple example, you can see that grafting was able to perfectly reproduce the glycan array screening data. At first glance it may not seem useful, but the initial 3D co-complex was actually one of four structures generated by docking to the crystal structure of the unliganded mAb. By grafting and comparing to known experimental array data, we observe that this co-complex matches the experimental array data.
To learn more about this project, refer to the publications.
Please note, the CFG v4.0 array contained errors in the reported structures, which we confirmed with the CFG. The excel file in this tutorial (but not the data on the CFG website) has been altered so that the correct structures are applied to ligands 158 and 159. See below for details:
Corrections or Clarifications Associated with CFG v4.0 Glycan Array Annotations
- When an anomeric center is not specified at the reducing end, the anomeric configuration is either not known or is present as a mixture.
- The sequence for ligand 158 (CFG v4.0 ID) is Galβ1-3(Galβ1-4GlcNAcβ1-6) GalNAc-Sp14. The anomeric center is undetermined, or is a mixture, the spacer is number 14.
- The sequence for ligand 159 is Galβ1-3(Galβ1-4GlcNAcβ1-6)GalNAcα–Sp8. The anomeric center is α, the spacer is 8.
- Ligands 157 and 159 are identical, Galβ1-3(Galβ1-4GlcNAcβ1-6)GalNAcα-Sp8.
- Ligands 125 and 182 are identical, Galβ1-3(GlcNAcβ1-6)GalNAcα-Sp8.