Here is an image of a typical page where glycosylation sites are listed. The “Show All” buttons have been clicked so that all sites are shown, and not merely the most biologically relevant.
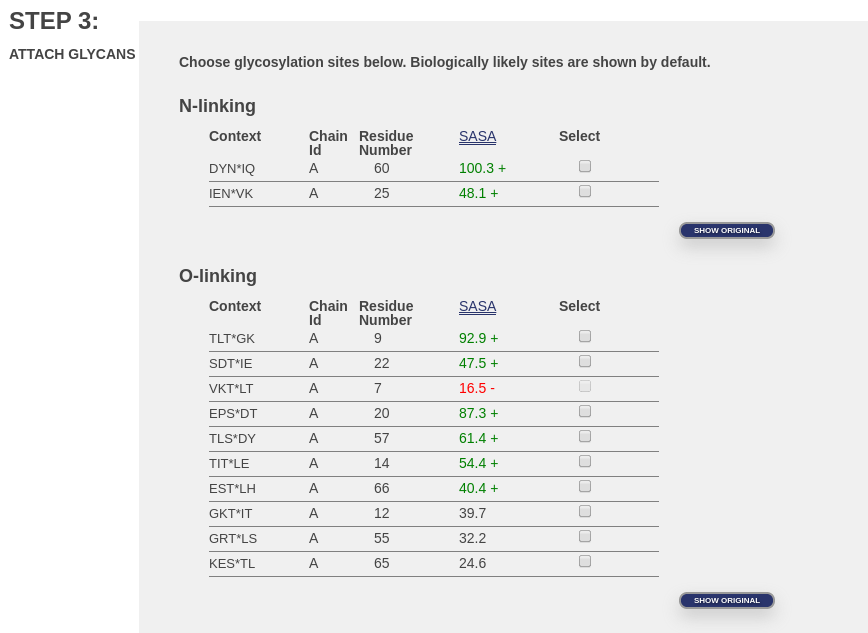
These are potential glycosylation sites in PDB ID 1UBQ.pdb. Notice that the third location under “O-linking”, for residue 7, is not available (the button is grayed out). That glycosylation site is unavailable because it is buried inside the protein. We can tell that it’s buried because we performed a calculation to determine the approximate amount of surface area of each amino acid that can be touched by a water molecule. This surface area is called the solvent accessible surface area or SASA (see column in image), and it is calculated by NACCESS. We know from experience that below a certain SASA value, it is very unlikely that glycosylation is possible. Even if the amino acid were oriented just perfectly to allow glycosylation, our attachment algorithm is not (yet) capable of such a careful attachment. SASA values that fall below this limit are given in red and marked with a minus sign (‘-‘). SASA values that indicate very exposed sites are given in green and marked with a plus sign (‘+’). Intermediate values are indicated by regular text with no marker.
If you really expect your site to be accessible, the problem might be, but is not necessarily, with your protein structure. Protein structures do not always represent their most biologically relevant conformation. For example, if the structure is from a crystal, the constraints of crystallization might have caused some part of the protein to close or collapse. It is also possible that the structure was obtained at a pH or salt concentration than is unlikely in vivo. That is, the conditions under which your structure was determined might have altered the structure so that a normally exposed site is not so exposed.
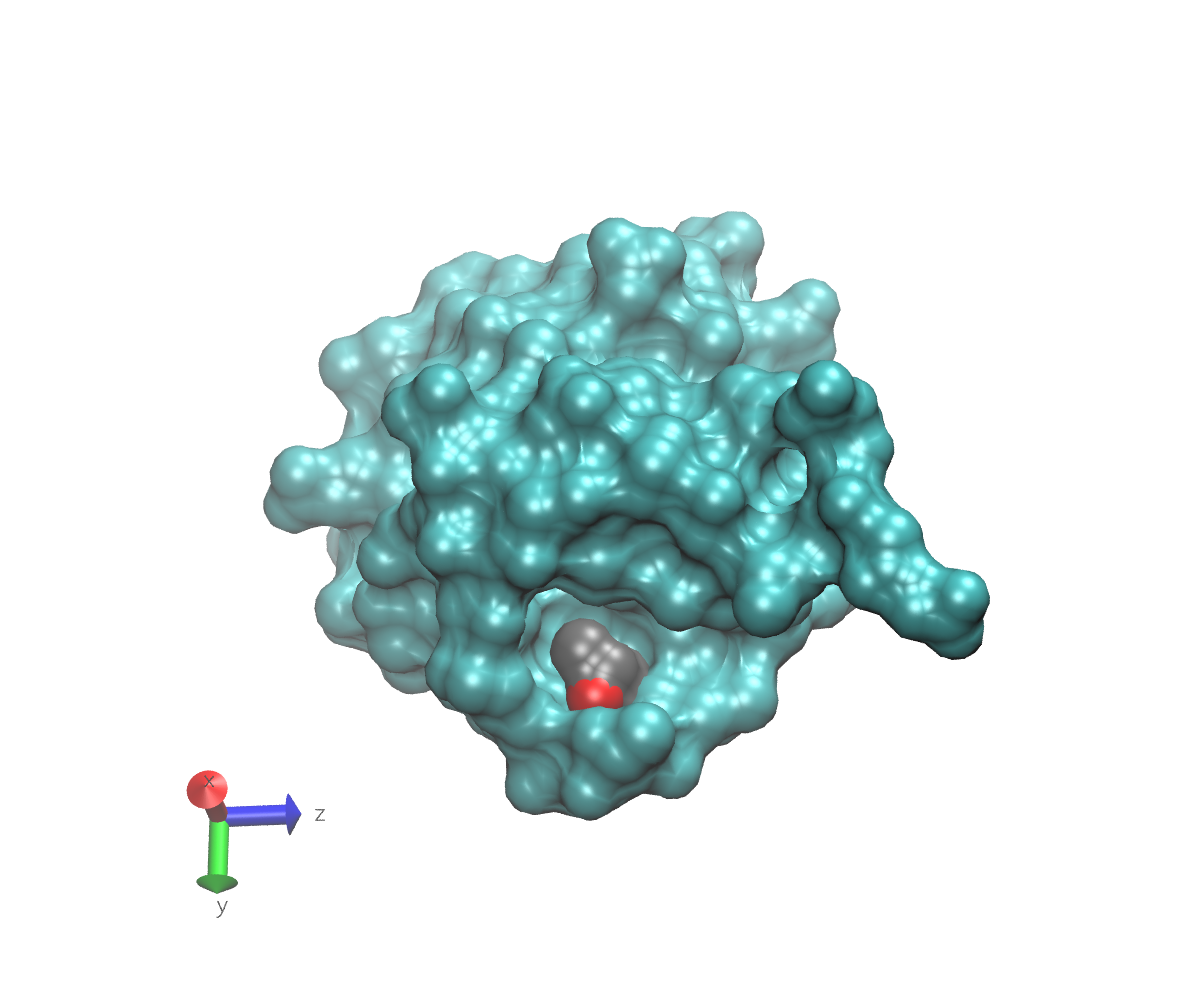
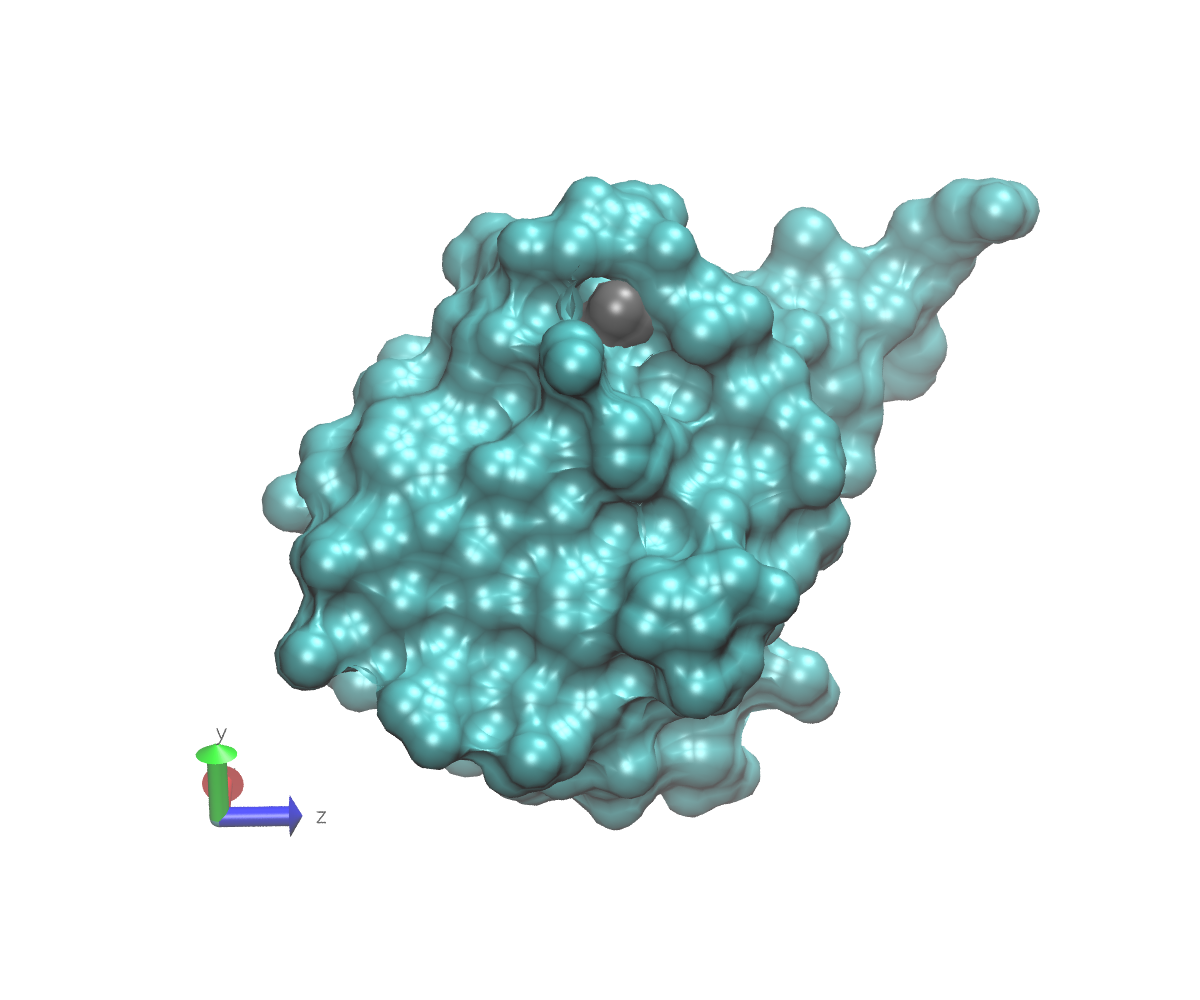
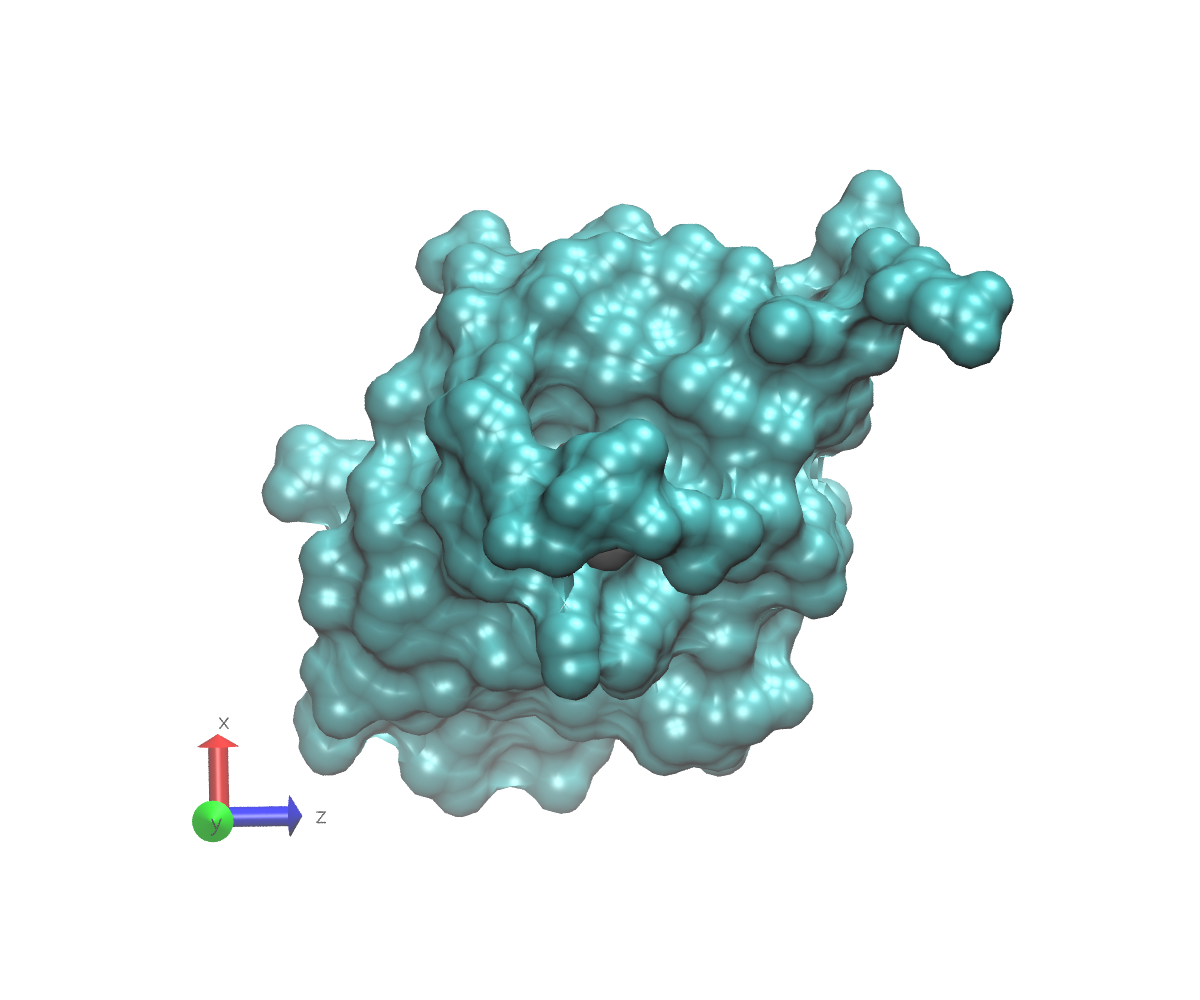
To get an idea what we mean by buried, here are some images of 1UBQ that emphasize the location of residue 7 relative to the rest of the protein. In each image, the protein surface except for residue 7 is colored cyan. The surface of residue 7 is colored gray. The atom to which a glycan might attach is represented as a red sphere that protrudes slightly from the gray surface. Note the axes in the lower left that can help you understand the orientations. These images were generated using VMD.
This first image shows the view from directly “above” residue 7. If you look carefully, you can just see a small patch of gray.
In this next view, the protein has been turned down and towards you (see axes). This is the most accessible that the linking oxygen is. Note that although some solvent can access it, the residue is inside a deep pocket.
This final view just shows the protein if it had been turned down and away from you. The residue is still visible, but obviously buried, and the linking oxygen is not visible at all.