Contents
Carbohydrates in the PDB and VMD
For reasons that are mostly historical, carbohydrates are currently not treated well in non-specialist databases such as the PDB or in visualization software such as VMD. Efforts are underway to improve the situation. In the meantime, this exercise will explain some of the problems, and offer some ways to deal with them.
The RCSB and the PDB
The Research Collaboratory for Structural Bioinformatics (RCSB, http://rcsb.org) maintains the Protein Data Bank (PDB), an extensive store of 3D structures of macromolecules of biological interest. The data are offered in a variety of formats, but the most commonly used is probably the PDB file. PDB files are popular because, while they are written to be read by a computer program, they are also easily readable by humans. 
Finding Carbohydrates in the PDB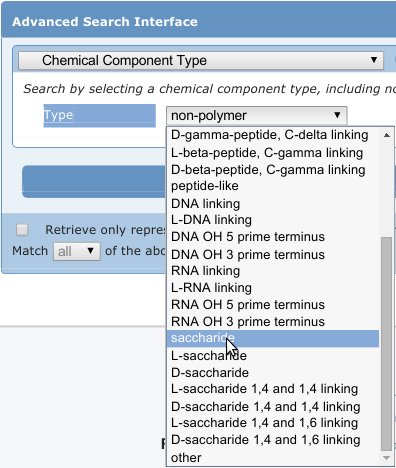
Currently, resources for identifying carbohydrates in the PDB are somewhat limited. For example, the search shown in the image at right (as of this writing) returns 3503 structures containing carbohydrates, with 195 unique ligands represented in those PDB structures. However, there are many more than that present. In addition to studies available in the literature, our recent, in-house survey found well over 11,000 PDB structures containing carbohydrates.
Here, we present three example structures containing carbohydrates. These are listed below, in order, from top to bottom, by decreasing ease with which you will be able to use VMD to visualize one of the glycans all by itself.
- 1G1Y: This is the structure of an alpha-amylase complexed with β-cyclodextrin, a ring made of seven α-d-glucose residues. The makers of Febreeze identify cyclodextrin as the ingredient that traps odor molecules so well.
- 1G1R: This structure contains sialyl LewisX (SLeX) bound to the lectin/EGF domain of P-selectin. SLeX is involved in cell-cell recognition and has an important role in human egg fertilization.
- 1RVZ: This structure of hemagglutinin from the 1934 flu is complexed the human pentasaccharide receptor. Only three of the five glycan residues could be resolved, so only three are visible in each of the 6 binding sites.
Downloading Structures from the PDB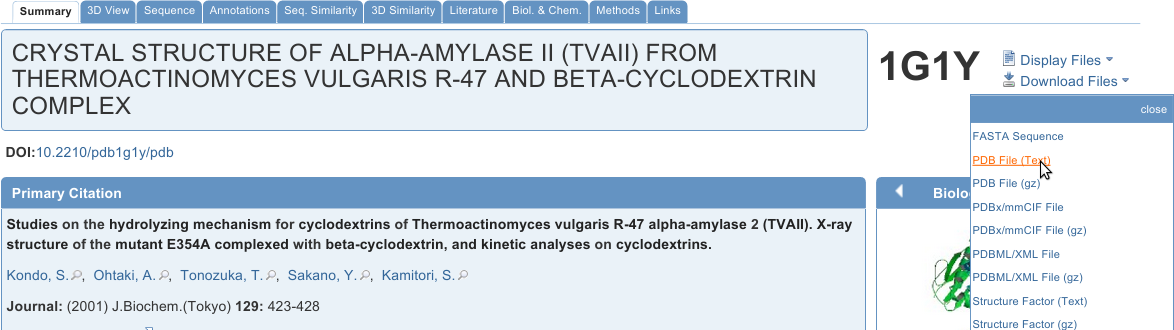
The simplest way to download a structure from the PDB is by typing the PDB ID into the search box shown above. Go to rcsb.org now and type 1G1Y (case insensitive) into the search box, then hit enter or click Go. Feel free to look around on the page, but also download the PDB File (Text) as shown in the image at right. Repeat the process for 1G1R, 1RVZ, and for any other file that is of interest to you.
Inspecting the Contents of PDB Files
Although the website for any given structure contains a wealth of information about it, a large  portion of that information is also contained within a PDB file. Open the file 1G1Y.pdb usingyour text editor. A sample image from gedit is shown at right. Other editors, including Notepad++ and TextWrangler, should have similar appearances.
portion of that information is also contained within a PDB file. Open the file 1G1Y.pdb usingyour text editor. A sample image from gedit is shown at right. Other editors, including Notepad++ and TextWrangler, should have similar appearances.
Look around in the file and see if you can answer the following questions. Hint, in many text editors, the key combination ctrl-f (both at the same time) will open a search dialog. The searchmight or might not be case-sensitive.
- What system is this a structure of?
- Is this structure from a native state, or one that has been changed? How?
- What sort of experiment was done to obtain this 3D representation of the system?
- What publication would you consult if you want to learn more?
- Find one or more ways in which this structure’s geometry (bond, angle or torsion) deviates from expected.
- What residue name is given to the cyclodextrin?

That last question illustrates one of the reasons that it is currently difficult to find carbohydrates in the PDB: it is not possible to perfectly predict how a carbohydrate will be encoded (named). In this case, the β-cyclodextrin (image, right) is encoded as a single entity rather than as seven α-d-glucopyranose residues linked 1-4 to each other in a ring. So, this structure would not be found from a search on glucose.
Visualizing the Structures Using VMD
First, we will focus on visualizing the protein – after all, the protein is likely to be an important part of any structure in the PDB. After that, we will explore representations of the carbohydrates.
In all structures shown here, we have set the background to white (Graphics -> Colors -> Display -> Background -> 8 White), the Display to Orthographic, and the axes have been turned off (Display -> Axes -> Off).
Visualizing the protein
- Open VMD, Go to File and choose New Molecule.
- In the Molecule File Browser, Browse to the location of 1G1Y.pdb and Load
- The viewing defaults are not ideal (see image at right). Try these changes:From Main, open Graphics -> Representations.
- In Selected Atoms, type protein.
- For Drawing Method, choose New Cartoon, and choose Secondary
 Structure for the Coloring Method (next image at right). Now, you should better see how the molecule is organized, but let’s do some more.
Structure for the Coloring Method (next image at right). Now, you should better see how the molecule is organized, but let’s do some more.
- It so happens that this structure is really two twin structures of the amylase bound to cyclodextrin. It also happens that each amylase is comprised of a single chain; so, we can easily view the symmetry.
- Now, let’s simplify things and add some visual interest.
- In Selected Atoms, add “and chain A” so that it reads “protein and chain A”. Hit enter.
- Click into the display window and type ‘=’ (the equals sign).
- Change the Coloring Method to Secondary Structure.
- Turn it around a bit, and you should see something like the structure just below right.
- For this step, first determine whether you have fast graphics or slow. If you don’t know, try the instructions assuming slow. If the response is very quick and smooth, try assuming fast. If the graphics become slow to respond when you rotate then in 3D, go back to slow.

- In Graphical Representations, click Create Rep to make a duplicate of your current representation.
- Change Drawing Method to Surf (for fast graphics) or QuickSurf.
- Surf is better than QuickSurf, but takes a lot more computation to draw.
- Change the Material to Transparent or Edgy Glass. Feel free to try others, but not all Materials render well on the computer screen without changes that require additional computation.
- Change the color of the surface, too, if you like. In the image at right, the chosen color is
 silver.
silver.
Do not close VMD or change any of your representations before moving on to the next part.
Visualizing the carbohydrates
For reasons similar to those that frustrate carbohydrate searches at the PDB, there are currently no easy ways to select carbohydrates in VMD. That is, you cannot enter “carbohydrate” as the Selected Atoms like you did with “protein”. However, with a little work, you can achieve acceptable results now.
- First, simplify your representation of chain A of the protein.Double-click on the surface representation (in Graphical Representations) to turn its display off.
- Change the Coloring Method for the New Cartoon representation to Color ID. Choose a color you like.
- You should now see something similar to the image (gray cartoon) at right.
- Now, show everything that isn’t protein.
- Click Create Rep in your Graphical Representations

- In Selected Atoms, type “not protein and chain A”.
- Change the Drawing Method to VDW and the Coloring Method to Name.
- You will now see all entities that are associated with chain A that are not protein. In particular, you should see many red spheres (image, right). These are water molecules trapped in the structure during crystallization.
- Now, change Selected Atoms to be “not protein and not water and chain A”. Now, the carbohydrate should be plainly visible (see image below right).

- Click Create Rep in your Graphical Representations
Sometimes, it will be harder to isolate the carbohydrate. Close VMD, reopen it, and load 1G1R.pdb.
- Open Graphical Representations. Change the existing representation to show “not water and not protein” (in Selected Atoms). Change the Coloring Method to Name, and set the Drawing Method to CPK, Licorice or VDW.
- You might notice that some entities are definitely not carbohydrates.
- Go VMD Main -> Mouse -> Label -> Atoms. Then, click on the three entities that are likely not to be carbohydrates. See the display at right. In this display, the label color was changed
 via Graphics -> Colors, and the size and weight of the labels was changed in Global Properties in the dialog opened by Graphics -> Labels.
via Graphics -> Colors, and the size and weight of the labels was changed in Global Properties in the dialog opened by Graphics -> Labels. - Now, you have some choices; here are a few. Try setting Selected Atoms to the suggestions below. Remember to type ‘=’ in the graphics window after each change so that the view resets to the currently visible atoms.
- not protein and not water and not resid 803 805 806
- not protein and not water and chain A and not resid 806
- not protein and not water and chain B
- not protein and not water and chain D (shown, licorice drawing, colored by Name). In the image shown (and likely in your representation), the strange bonds leading from the fucose (see label in image) indicate that it is complexed to the adjacent calcium ion.

Of course, you could also determine the residue names or IDs for the carbohydrates and list them as the selected atoms to display.
Now, load 1RVZ.pdb. Set “protein” to display as a single-color cartoon, and set “not protein and not water” to display as VDW, colored by Name. See the image below. In the image below, the cartoon representation has been modeled with the Edgy Glass Material to make the carbohydrates more visible. Isolating this file’s carbohydrates for visualization is certainly possible, but requires a bit more work than with the previous files.
If there is extra time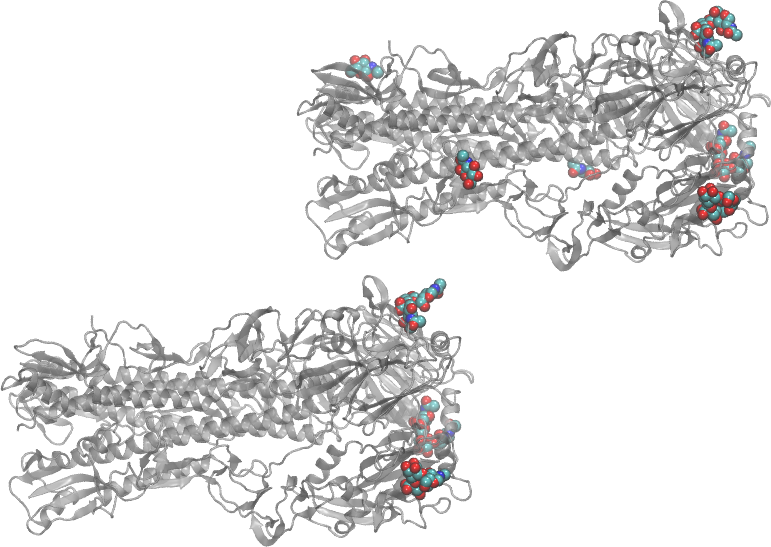
Load 1RVZ.pdb and display one or more of the mono- or oligosaccharides individually. Alternately, find another carbohydrate-containing PDB file that is of interest to you and display the carbohydrates.

